Eleanor Hall, Gut Reaction Programme Manager, discusses our ongoing work to define an acceptable and sustainable model for health data research, and the importance of our new Cohort Discovery Tool in this process
Health data is a key resource for driving clinical innovation, but to fully realise its potential we must meet the challenge of maintaining secure, transparent and appropriate use of participant information. The Health Data Research UK (HDR UK) Innovation Gateway allows researchers to discover available datasets and to understand high-level information about what is present, permitting potential users to plan their data request (Figure 1, HDR UK Innovation Gateway). The eleven Gut Reaction datasets currently listed on the Gateway contain information on the fields available, levels of data completeness, overall volumes, and how to apply for data access. While it is useful to know that a dataset exists and what it contains, ongoing engagement with our industry partners confirms that some research planning requires more detailed knowledge of the underlying data – for example whether there are sufficient records of linked data to satisfy a particular set of conditions to make a project feasible.
Under our previous model, the Gut Reaction data team ran these queries on behalf of the researcher; this often involved a back-and-forth conversation to find the optimal query with sufficient datasets for the research question to be viable. Feedback from commercial companies tells us that the ability to self-adjust queries and receive instant results would speed up decision-making about access applications to the Gut Reaction datasets, removing a hurdle that slows down crucial research.

Figure 1: the HDR UK Innovation Gateway
In order to maximise the secure use of data, and allow researchers greater confidence in the feasibility of their studies prior to a full data request, software enabling users to safely but accurately determine how many records meet a particular set of criteria has been developed. Our Cohort Discovery Tool employs open source i2b2 technology allowing researchers to directly query de-identified, aggregated patient datasets gathered from multiple sources. The Gut Reaction team coded the data within the Cohort Discovery Tool using the NHS data standard, SNOMED-CT, to allow searches across multiple datasets. Two Gut Reaction datasets, held by the NIHR IBD BioResource, have been made available in this way; Case Report Forms (CRFs), phenotypic data collated and recorded by Specialist Research Nurses during enrolment in the NIHR IBD BioResource, and lifestyle questionnaire data provided by the participants themselves, with other datasets to follow.
Working with our Patient Advisory Committee, this tool aims to balance researcher needs and the requirement for data privacy in a way that works for all of our stakeholders. These discussions confirmed the absolute necessity for all data to remain de-identified1, and Gut Reaction has taken several steps to ensure this. Our approach includes using the Privitar privacy enhancing software, alongside removing directly identifying data (such as names and postcodes) and placing some precise data (including date of birth, height and weight) into broad groups. Search criteria resulting in very low counts, such as unusual combinations of medications, could theoretically risk the possibility of an individual being identified, so these are hidden within the Cohort Discovery Tool, and full data access request approval is required for additional details in these cases.
Crucially, dataset security is assured by only allowing approved researchers access to the Cohort Discovery Tool through our Trustworthy Research Environment (TRE). The AIMES TRE ensures that once a cohort of patients has been selected, users will only see the number of records that meet their selected criteria; drainage of intra-abdominal abscesses in the image below (Figure 2). The underlying data is not visible to the user, and the TRE ‘airlock’ ensures that any results from using the Cohort Discovery Tool cannot be exported by the user unless, or until, a full data access application is received and approved. When a full data access application from the researcher is approved, the Gut Reaction team are able to export the results to identify the exact datasets required. This ensures that patient privacy is maintained.
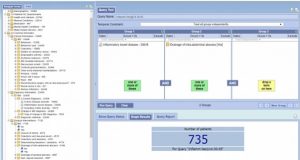
Figure 2: screenshot of the Gut Reaction cohort builder in use, to illustrate the fields available, the query builder, and outputs as a ‘cohort count’
Successful use of health data for research requires finding an acceptable balance between data safety and privacy, and data access. We are committed to working collaboratively with all of our partners to find the best way to do this; our Cohort Discovery Tool marks the next stage in this journey.
- For discussion on terms relating to anonymity see Understanding Patient Data


